Phenotyping the Tumor Microenvironment Using DNA Barcoded Multiplex Technology

Understanding the tumor microenvironment (TME) is essential for the development of effective treatments for cancer patients. Traditional multiplex IHC methods offer limited understanding of the complex TME. This is partially due to technical limitations of chromogenic dyes and fluorescent detection systems. Ultivue DNA barcode labeling technology (InSituPlex) provides an innovative way to detect multiple markers simultaneously in the same tissue section using an automated workflow on the Lecia Biosystems Bond RX platform. This webinar will share our firsthand experience with a two-step 7 marker assay and feature some of the extracted data.
Learning Objectives
- Understand the complexity of tumor microenvironment and its importance in drug development
- Identify the Ultivue DNA barcoding and exchange technology for multiplex immunofluorescence
- Examine the processes of image fusion, spatial analysis and colocalization of markers on individual cells
Webinar Transcription
- Phenotyping Tumor Microenvironment Using DNA Barcoded Multiplex Technology Chifei Sun GSK IVIVTBIBCellular Biomarkers Collegeville, PA
- I have no conflict of interests with Leica Biosystems, Ultivue and Indica Labs. – All the materials used in this presentation are for educational purpose only. – The human biological samples were sourced ethically, and their research use was in accord with the terms of the informed consents under an IRB/EC approved protocol. Statement 9/25/2020 2
- Understand the complexity of tumor microenvironment and its importance in drug development – Introduce the Ultivue DNA barcoding and exchange technology for multiplex immunofluorescence – Highlight the processes of image fusion, spatial analysis and colocalization of markers on individual cells Learning objectives Insert your date / confidentiality text here16x9 core template 3
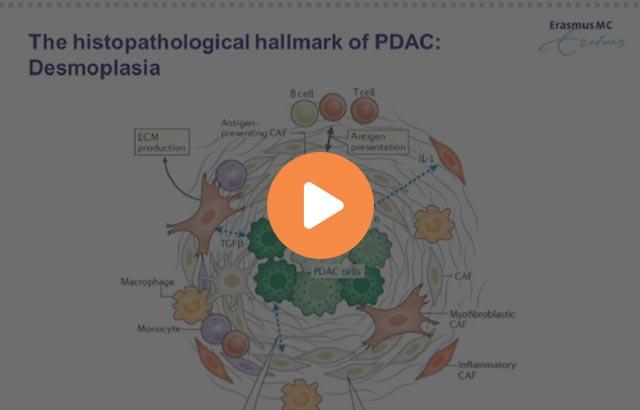
- Tumor microenvironment and cellular infiltration Sid P. Kerkar, and Nicholas P. Restifo Cancer Res 2012;72:3125-3130 Landscape of immunotherapy Galon et al. Nat Rev Drug Discov 18, 197–218 (2019)
- Biomarkers Improve Decision-Making Across R&D T2L L2C C2FTIH FTIH2POC POC2 File Target Acceptance: Agree Indication(s) Outline clinical plan Translational models Define Biomarker Strategy Candidate Selection: Biomarkers must be defined for FTIH/POC; initiate assay development clinical biomarkers FTIH/POC: Implement biomarkers and monitor clinical performance-review any gaps as they appear Phase IIb and III: Define biomarkers for inclusion in late stage studies and filing Define strategies for PD, surrogate and clinical biomarkers Use biomarker data from ex- vivo/preclinical models to model human dosing Confirm engagement of mechanism (PD) and assess clinical efficacy; test predictive biomarkers Confirm requirement for predictive biomarker: define efficacy endpoints Post Approval Phase IIIb/IV: Define biomarkers to support mechanism and Indication expansion Further refine populations and support new indications 5 – a measurable substance in an organism whose presence is indicative of some phenomenon such as disease, infection, or environmental exposure.
- Technology Overview – Why multiplex – Current practice involves the optimization of assays for single plex IHC using chromogenic dyes. These are used because they can evaluate using a standard brightfield microscope and morphological details are present to aid the interpretation. – Limited amount of patient tissue obtained from core needle biopsies – It facilitates the demonstration of co-localized markers and their measurement using image analysis. Single cell analysis. – Supports our movement to digital workflow and multiplex image analysis – Reduce stock of consumables and reduce waste due to discarding expired reagents Situation Drivers for change
- Technology Overview – Why multiplex – Select panel of HRP dyes that are acceptable for single plex IHC on the Ventana and Lecia Bond instruments. – Evaluate best practice by testing various approaches and then establish guidelines for selecting the optimal multiplex IF approach. – Here is a list of some of the available options: – Ventana IF 5-plex – HRP Tyramide – Opal detection – HRP Tyramide – Cell IDX – proprietary modified hapten based technology – Ultivue – DNA barcode labels, focus of this presentation Path Forward
- Ultivue Method - Points to Consider High specificity since each antibody is tagged with a unique DNA barcode linker. All primary antibodies are cock tailed and applied in a single solution and incubation step. This is a non-species dependent detection method so no concern about selecting and quenching or denaturing anti-species 2nd Ab. Tyramide and HRP free detection Signal is bright due to amplification method Protocol is fully automated and less than 6 hours.
- ULTIVUE INC. | DEC-2018 | PROPRIETARY - NOT FOR REDISTRIBUTION 9 • Dewax and retrieve samples as usual • Incubate sample with a mixture of barcoded antibodies in a single staining step • Unique DNA sequence (barcode) attached to each antibody • Add complementary fluorescent DNA probes to bind targets • Samples are ready to be imaged • DNA hybridization technology • Amplify all targets simultaneously • Increases ratio of barcodes per antibody allowing for more complementary probe strands to bind Amplification Detection Staining Single Antigen Retrieval Step Single Staining Step Single Amplification Step Single Detection Step Mixture of DNA- barcoded antibodies Mixture of fluorescent probes Assay Overview ➤ Ultivue, Inc.
- Workflow for standard 4-plex followed by H&E on same slide Automated Staining Whole Slide Imaging Coverslip Removal and H&E Whole Slide Imaging Coregister & Fusion Image analysis Imaging systems require Cy7 Filter Automated Staining (BOND RX - Leica) WS Fluorescent Scanner (Vectra Polaris) User dependent Software (HALO – Indica Labs) Same day Sample-to-Images WS Brightfield Scanner (Vectra Polaris) H&E Stain (HE 600)
- Two ready to use panels were selected from Ultivue for testing on our Leica Bond RX. UltiMapper I/O PD-1 Antibody Panel UltiMapper I/O PD-L1 Antibody Panel Antibody Conjugate Clone ID Detection Channel Cell Type CD45RO UCHL1 FTIC Memory T- cells PD-1 CAL20 TRITC Exhausted or suppressive T-cells CD3 BC33 Cy5 T-cells panCK/SOX10 Cocktail AE1/AE3 /BC34 Cy7 Tumor cells Epithelium Antibody Conjugate Clone ID Detection Channel Cell Type CD8 C8/144B FTIC Cytotoxic T- cells CD68 KP-1 TRITC macrophages PD-L1 73-10 Cy5 Immune Checkpoint Marker panCK/SOX10 Cocktail AE1/AE3/ BC34 Cy7 Tumor cells Epithelium
- Lecia Bond RX – Ultivue Protocol Standard bake and dewax followed by epitope retrieval 2 for 20 minutes. Solutions were made according to vendor recommendations using the reagent calculation worksheet. Reagents were loaded into titration containers. Reagent Temperature Time UltiMapper™ Ab Diluent Ambient 15 minutes Primary Ab Cocktail Ambient 60 minutes Bond Wash Buffer Ambient 3 X 0 minutes UltiMapper™ Pre-Amplification Mix Ambient 25 minutes Bond Wash Buffer Ambient 35°C 3 X 0 minutes 1 X 5 minutes UltiMapper™ Amplification Solution Ambient 90 minutes Bond Wash Buffer Ambient 3 X 0 minutes UltiMapper™ Nuclear Counterstain (Hoechst) Ambient 1 X 0 minutes 1 X 15 minutes Bond Wash Buffer Ambient 3 X 0 minutes UltiMapper™ Fluorescent Probes Ambient 2 X 12 minutes Bond Wash Buffer Ambient 2 X 0 minutes 1 X 20 minutes
- Tonsil: PD-L1 Panel Tonsil: PD-1 Panel Vectra Polaris Images: Multiplex IF and Reference H&E
- Stomach tumor core: PD-1 Panel with panCK/SOX10 Stomach tumor core: PD-L1 Panel with panCK/SOX10
- PD-1 Panel CD45RO PD-1 CD3 panCK/SOX10 PD-L1 Panel CD8 CD68 PD-L1 panCK/SOX10
- Custom 7-plex Immunophenotyping panel Case Study
- ULTIVUE INC. | DEC-2018 | PROPRIETARY - NOT FOR REDISTRIBUTION 18 ➤ High level of multiplex ➤ No signal unmixing ➤ Single slide, whole slide imaging ➤ Single day, sample to data ➤ High throughput Workflow of an 8-plex on a Single Slide, Dewax and retrieve sample Incubate sample with all antibody-barcode conjugates in a single staining step Amplify all barcodes simultaneously Add 1st set of fluorescent probes to bind targets Image sample Remove 1st set of probes Add 2nd set of fluorescent probes to bind different targets Image sample Assay Overview ➤ Ultivue, Inc.
- Ultivue 7-plex phenotyping and expression markers Fit-for-purpose optimization of multiplex IHC assays UltiVue Assay Panel kit MTO 7-plex Panel Description Target Role IHC Target Markers Nuclear Counterstain Round 1 Round 2 CD8, CD68, PD-L1 Ki67, GZMB, CD3, PanCK Phenotyping CD3 CD8 Hoechst CD68 Expression Granzyme B (GZMB) Ki67 PD-L1 Segmentation PanCK
- Phenotyping goals/priority table Fit-for-purpose validation of multiplex IHC assay Cell Types Reporting Ranking for prioritization of analytical validation Tissue Segments Parameters Phenotypes 1. CD3+/CD8+ 2. CD68+ 3. GZMB+ 4. PanCK+ 5. PanCK+/Ki-67+ 6. CD3+/CD8+/Ki-67+ 7. PanCK+/PD-L1+ 8. CD3+/CD8+/PD-L1+ 9. CD68+/PD-L1+ 10. CD68+/PD-L1- 11. CD3+/CD8+/Ki-67- 12. CD3+/CD8+/PD-L1- Tumor Non-Tumor Total Count Cell Density (cells/mm2) Percentage of all cells Summary Total Cells
- These are representative images of CD3 staining (DAB-brown) to classify Hot & Cold tumors – Hot tumors respond to checkpoint inhibitors – Cold tumors can be turned into Hot (epigenetic drugs, chemo, rad, etc.) – Hot/Cold classification could be based in several markers: – T-cell – myeloid markers, – proliferative index, – mutational burden Implications of HOT/COLD classification in patient outcome Why we want to understand the tumor microenvironment (TME) 16x9 core template 21 Galon et. al, 2019 CD3
- Rep. images “Hot” immune inflamed tumor tissues
- CD8 PanCK Sample G20-0002 (bladder)
- CD8 PanCK CD3 CD68 Ki67 PD-L1 GzmB Sample G20-0002 (bladder)
- Rep. images “Excluded” immune-excluded tumor tissues
- CD8 PanCK Sample G20-0008 (bladder)
- CD8 PanCK CD3 CD68 Ki67 PD-L1 GzmB Hoechst Sample G20-0008 (bladder)
- Rep. images “Cold” immune-dessert tumor tissues Chifei Sun& Fang Xie BIB, IVIVT
- CD8 PanCK Sample G20-0003 (bladder)
- Sample G20-0003 (bladder) CD8 PanCK CD3 CD68 Ki67 PD-L1 GzmB
- Sample G20-0005 (bladder) CD8 PanCK CD3 CD68 Ki67 PD-L1 GzmB
- Insert your date / confidentiality text here16x9 core template 32 Ultivue 7- plex – NSCLC; selected combinations CD3 CD8 CD3 CD68 CD3 GzmB PanCK CD68 PD-L1 PanCK CD3 Ki67 PanCK CD3 CD68 PanCK CD3 PD-L1 PanCK CD8 CD68
- Bladder Tumors Classification as “Cold, Hot or Excluded” by pathologist 33
- Assay Workflow 1. Multiplex immunophenotyping staining on the Leica Bond RX 2. Qualitative assessment and classification of TME as inflamed (HOT), immune-excluded (EXCLUDED) or immune desert (COLD); visual analysis by pathologist 3. Step for HALO Image analysis of immunophenotypes • Serial staining images fusion • Tumor/stroma classification • Cell detection and phenotyping (PanCK+, CD3+CD8+, CD68+ and GzmB+ cells)
- 35 Image Analysis workflow Staining Round 1 Staining Round 2 Fused image Classification Tumor/stroma Cell detection Phenotyping Infiltration spatial plot 1. Fuse images 2. Run PanCK classifier algorithm (Random Forest AI) 3. Run cell detection algorithm (Hoechst ) 4. Phenotyping ( IHC -fluorescence). Phenotype populations counted for epi. Tumor, non tumor (stroma) 5. Infiltration study & spatial plots Software: HALO v3.0
- www.indicalab.com Analyze an unlimited number of markers in any compartment in fluorescence. Any cell compartment can be analysed – membrane, cytoplasm, nucleus. Reports user-defined cell phenotypes based on biomarker expression. Fluorescent analysis with HALO ® Highplex FL
- www.indicalab.comwww.indicalab.com High-Plex Immunofluorescence Images Nuclei CD FoxP3 CD8 CD68 CK CD163 Phenotypes defined through combinations of marker/channel positivity Results viewed for entire panel or specific phenotypes Immune-phenotype Understanding the immune-phenotype within the context of the tumor microenvironment Hoec hst CD4 CD8 FoxP3 PD-1 Mel A
- Hoechst cell segmentation seems to fuse some nuclei in dense areas, a possible solution is to increase “segmentation aggressiveness” in HALO IA – No more than 10% of cells are lost by this “fusion” segmentation artifact – Disadvantage of increasing “segmentation aggressiveness” is that the algorithm will segment nuclei of big tumoral cells; historical observations – CD3-IA is sensitive and picks up >90% of the cells – CD8-IA seems to be missing some low expressing cells (10-20%); we suggest a lower threshold for future analysis using this algorithm – CD68-IA is sensitive and picks up ~90% of the cells – GZMB-IA was difficult to evaluate in samples where <1,000 positive cells were detected; we suggest for future studies that a strong positive control is included – 4 samples were excluded from IA because of excessive autofluorescence or image misalignment General notes Image Analysis (IA) QC, visual assessment 38
- Quant. Analysis of Bladder Cancer Insert your date / confidentiality text here16x9 core template 39 PanC K + C ells C D 68+ C ells C D 3+/C D 8+ C ells G ZM B + C ells 0 500 1000 1500 2000 2000 4000 6000 8000 10000 Phenotype Analysis, Bladder Cancer (N=16) cell counts/mm2 Cells/mm2 Total Tumor Non-tumor PanC K + C ells C D 68+ C ells C D 3+/C D 8+ C ells G ZM B + C ells 0 5 10 15 20 20 40 60 80 100 120 Phenotype Analysis, Bladder Cancer (N=16) percentage of total cell population PercentofDAPI+cells Total Tumor Non-tumor
- Quant. Analysis of Bladder Cancer Insert your date / confidentiality text here16x9 core template 40 C D 3+/C D 8+/K i-67+ C ells C D 3+/C D 8+/K i-67-C ells C D 3+/C D 8+/PD -L1+ C ells C D 3+/C D 8+/PD -L1-C ells C D 68+/PD -L1+ C ells C D 68+/PD -L1-C ells PanC K +/K i-67+ C ells PanC K +/PD -L1+ C ells 0 5 10 15 20 20 40 60 80 100 Phenotype Analysis, Bladder Cancer (N=16) percentage of main phenotype Percentage Total Tumor Non-tumor C D 3+/C D 8+/K i-67+ C ells C D 3+/C D 8+/K i-67-C ells C D 3+/C D 8+/PD -L1+ C ells C D 3+/C D 8+/PD -L1-C ells C D 68+/PD -L1+ C ells C D 68+/PD -L1-C ells PanC K +/K i-67+ C ells PanC K +/PD -L1+ C ells 0 200 400 600 800 Phenotype Analysis, Bladder Cancer (N=16) cell counts/mm2 Cells/mm2 Total Tumor Non-tumor
- Bladder cancer, hot/cold plot HALO result in good agreement with path evaluation Insert your date / confidentiality text here16x9 core template 41 Bladder Tumors Visual Classification as “Cold, Hot or Excluded” by pathologist G 20-0002G 20-0003G 20-0004G 20-0005G 20-0006G 20-0007G 20-0009G 20-0010G 20-0011G 20-0013G 20-0014G 20-0015G 20-0016G 20-0018G 20-0019G 20-0020 0 10 20 30 40 50 60 70 Bladder Cancer, N=16 Percentage of TILs (CD3+/CD8+, CD68+, GZMB+) PercentofDAPI+cells Tumor Non-tumor hot excl. Cold
- Spatial plot CD3+/CD8+ distribution in bladder tumor Insert your date / confidentiality text here16x9 core template
- Proximity analysis Measure the distance of each cytotoxic T cell to the nearest tumor cell Insert your date / confidentiality text here16x9 core template
- Nearest neighbor analysis Find the nearest cytotoxic T cell for each tumor cell Insert your date / confidentiality text here16x9 core template 44
- Infiltration analysis Count the number of cytotoxic T cells in tumor and stroma Insert your date / confidentiality text here16x9 core template 45
- Acknowledgements BIB Cellular Biomarkers Clinical Histotechnology Bao Hoang Haydee Lara Chifei Sun Fang Xie David Krull US Pathology: Leslie Obert Daniela Ennulat Indica Labs Ultivue
About the presenter

Chifei is a senior histotechnologist with GSK R&D. He has broad knowledge and technical skills in routine histology, immunohistochemistry, multiplex immunofluorescence, digital pathology and quantitative image analysis. His work scopes from target validation, animal model characterization to clinical biomarker assay development and validation. He has a master’s degree in neurobiology from Fudan University and a bachelor’s degree in biology from Shandong University in China.
Related Content
Die Inhalte des Knowledge Pathway von Leica Biosystems unterliegen den Nutzungsbedingungen der Website von Leica Biosystems, die hier eingesehen werden können: Rechtlicher Hinweis. Der Inhalt, einschließlich der Webinare, Schulungspräsentationen und ähnlicher Materialien, soll allgemeine Informationen zu bestimmten Themen liefern, die für medizinische Fachkräfte von Interesse sind. Er soll explizit nicht der medizinischen, behördlichen oder rechtlichen Beratung dienen und kann diese auch nicht ersetzen. Die Ansichten und Meinungen, die in Inhalten Dritter zum Ausdruck gebracht werden, spiegeln die persönlichen Auffassungen der Sprecher/Autoren wider und decken sich nicht notwendigerweise mit denen von Leica Biosystems, seinen Mitarbeitern oder Vertretern. Jegliche in den Inhalten enthaltene Links, die auf Quellen oder Inhalte Dritter verweisen, werden lediglich aus Gründen Ihrer Annehmlichkeit zur Verfügung gestellt.
Vor dem Gebrauch sollten die Produktinformationen, Beilagen und Bedienungsanleitungen der jeweiligen Medikamente und Geräte konsultiert werden.
Copyright © 2024 Leica Biosystems division of Leica Microsystems, Inc. and its Leica Biosystems affiliates. All rights reserved. LEICA and the Leica Logo are registered trademarks of Leica Microsystems IR GmbH.