
Spatial Biology


Several tools within the landscape of scientific disciplines accurately quantify or locate a molecule of interest. These tools include immunohistochemistry (IHC); in situ hybridization (ISH); single-cell RNA sequencing for protein, DNA, or RNA detection; and several other techniques to identify a target’s molecular expression. However, these techniques have been constrained by a lack of understanding of the entirety of the neighboring cellular environment. Spatial Biology is a technology that uses fluorescent imaging and other tools to analyze molecular expression at the single-cell and subcellular levels, in order to understand that expression among other cells within a tissue. This article gives an overview of the importance of Spatial Biology, its use in multiomics, and technologies using spatial analysis (for example, multiplex IHC, multiplex immunofluorescence, or flow cytometry). We also discuss the role of artificial intelligence (AI) and the future of Spatial Biology in advancing human health.
1. Why is Spatial Biology important?
Spatial Biology is the 2–3-dimensional study of expression of genes and proteins and their corresponding interaction with the cellular environment. By analyzing real-time molecular behavior of multiple markers, spatial biologists can begin to piece together a larger picture of cellular and tissue function.1 disease, In the past, the study of cellular morphology relied on qualitative microscopy, while molecular processes and signaling pathways were studied by isolating tissue and studying cellular interactions in a silo via genomics or transcriptomics .2 However, with the advent of spatial biology, researchers are now able to explore interactions in situ – in their physiological location.2 Spatial transcriptomics and proteomics are steadily finding use in deducing vital information, including single cell transcriptomes, single cell spatial protein expression and interaction,3 and comparisons of RNA and protein in the tumor microenvironment. For example, the developmental mechanism of a multicellular organism from a single totipotent (stem) cell is better understood now than ever before, thanks to spatially deciphered relationships of complex cellular interactions.4 Similarly, studies highlighting the subclonal heterogeneity of the tumor microenvironment have given scientists important information about disease management via spatial research.
While techniques such as flow cytometry and single cell sequencing also provide information on the single cell transcriptome or proteome, these tools cannot evaluate the in situ spatial environment.
Immunohistochemistry, on the other hand, delivers information about protein localization but cannot provide information on a protein’s interaction with neighboring molecules. Spatial biology allows researchers to discover new information about the complex interactions of DNA/RNA/proteins in a spatiotemporal setting by utilizing single cell sequencing in combination with IHC/IF and other such tools, to ultimately provide a holistic idea of the dynamics between a cell and its neighboring components.
2. Multi-omics and Spatial Biology
2.1 Spatial transcriptomics
Single-cell RNA sequencing (scRNAseq) was developed in the early 2000s, and it has aided in analyzing the subcellular nature of mRNA, for example, its structure at the time of synthesis and the location where it is trafficked.5 This has given researchers a method of evaluating total RNA and its function across disease contexts, in what is called spatial transcriptomics.
Although still in their infancy, technologies are being developed in which a carefully designed probe binds to an RNA target and helps to identify the RNA location in its spatial environment. This complex technique is called single molecule fluorescent in situ hybridization (smFISH). Several FISH probes can simultaneously hybridize to corresponding RNA transcripts in real time to decode complex cellular interactions.
FISH-assisted transcriptomic limitations include spectral overlap where the fluorophore crowding can produce false results. To overcome this barrier, combinatorial spectral barcoding utilizes several fluorophores operating in unique spectra, which may prevent the uncertainty of accurate molecular identity. In addition, MERFISH, or multiplex error FISH, is an updated version of smFISH, which aids in identifying thousands of RNA species in a sequentially manner.6
2.2. Spatial proteomics
More than 10,000 proteins are expressed in a cell, and the expression profile of these proteins (the proteome) is unique in every cell. Subcellular localization, post-translation protein modification, protein splicing, receptor endocytosis, and cellular environment, including metabolic changes, are some ways a protein is uniquely expressed and interacts with its counterparts. Post-translational activity is dynamic and can be tracked through spatiotemporal analysis. 7 Several methods have been developed to analyze the spatial biology of a cell.
Mass spectroscopy (MS) is a technique that aids in the spatial localization of the protein of interest. Through MS, organelles can be fractionated, and apeptide profile can be visualized.7 Additionally, post-translational modifications can also be captured through this tool. However, MS has some drawbacks, including inability to accurately identify single cell content. With MSI (mass spectroscopy imaging), by analyzing molecules’ charge/mass (z/m) ratio through laser assisted ionization, heat maps are generated that help in the positional evaluation of the entire proteome.8
IF-based spatial visualization of proteins is another tool for localization analysis, utilizing corresponding antibodies against individual protein targets. More than 2,000 proteins have been located within a single tissue section by measuring fluorescently tagged proteins. Separately, by inserting a fluorescently labeled gene into the cellular genome, the life cycle of the corresponding coded protein can be tracked to monitor its morphological and physiological journey.
2.3. Integration of spatial transcriptomics and proteomics provides a systemic overview of cellular components
Spatial transcriptomics and proteomics can provide vital information related to specific biomarkers. Integration of these two techniques can provide important information on cell-cell interaction, which enables the scientific community to understand complex genomic and proteomic tissue structure.9 Below is the difference between the two technologies.
Table 1. Comparison of spatial transcriptomics and proteomics
| Spatial transcriptomics | Spatial proteomics |
|---|---|
| Can identify gene expression by detecting transcript expression at a bulk and single cell level | Can detect protein, splice variants, and other post-translational modifications in a single or bulk cell population |
| Can detect and sequence thousands of transcript/genes simultaneously | Can detect tens of proteins simultaneously |
| Pivotal for fundamental research | Proteome-based data can drive clinical research in developing viable treatments for patients |
3. Methods Adapting Spatial Biology
3.1. Multiplex Immunohistochemistry
Multiplex IHC analyzes several biomarkers within one tissue section for improved understanding of protein localization and implications for disease management. Using chromogenic multiplex IHC, up to 5-7 dyes can be used to identify relevant biomarkers (Figure 1). Spatial knowledge of several biomarkers or proteins of interest gives more information on the disease state, its progression, and corresponding treatment.10 Techniques such as flow cytometry and single-cell RNA sequencing are essential tools to extract information about cellular genetic makeup; however, these techniques do not provide information on the cellular environment/localization. Traditional single or duplex IHC, similarly, may also give information on the molecular localization of 1 or 2 proteins; however, it does not provide a holistic view of complex cell interactions for a better physiological understanding of the disease state. Multiplex IHC has recently evolved to be at the frontier of spatial research. Through this tool, tens of different biomarkers can be identified.11
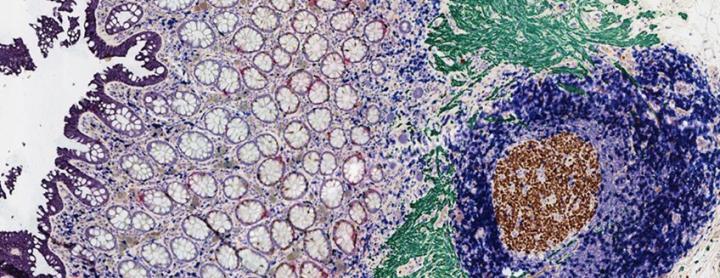
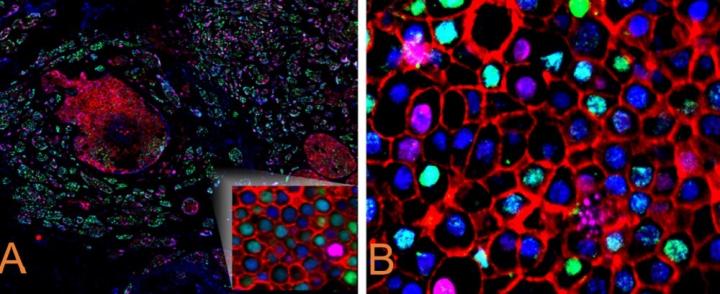
3.2. Immunofluorescence
Like multiplex IHC, multiplex immunofluorescence (mIF) is a valuable tool for identifying and detecting complex protein interactions. In most available technologies, mIF involves using several antibodies at one time raised against specific proteins of interest, and often using a cycling method to continually add more markers for simultaneous analysis within one tissue (representative image depicted in Figure 2). Because so many antibodies are used, and secondary antibodies are species-specific, it is usually essential to validate the robustness of both primary and secondary antibodies before using them to address a problem or hypothesis in mIF. Caution must be taken not to choose antibodies that are raised in the same species, as this may compromise the antibody specificity. In addition, each fluorophore emission and excitation wavelength must be noted to avoid background signal and spectral mixing.12
FOR RESEARCH USE ONLY. Not for Use in Diagnostic Procedures.
3.3. Optical Barcoding
The uniquely colored labeled RNA target molecule whose sequence corresponds to the gene of interest binds to the single-stranded unique reporter DNA and capture probe, which helps identify and measure the fluorescently labeled RNA transcriptional scripts of the gene of interest. Molecular DNA barcodes can locate the corresponding RNA and provide spatial information on the complex cellular environment.13
3.4. Mass spectrometry
Mass spectrometry is a technique that analyzes metabolic processes in an organism by leveraging the mass and charge of specific analytes. With the advent of spatial biology, mass spectrometry images have been reported to simultaneously identify more than 2000 biomolecules of the order of ~100 µm.14 Affinity purification-based MS is used to purify/isolate a specific protein and its binding partner, which is subsequently isolated by performing gentle chemical processing. A bait-based MS method is used to map out the cellular proteome. This highly resolved organismal proteome information can be translated for clinical use, albeit with further well-designed studies.
3.5. Cytometry
Fluorescence activated cell sorting (FACS) identifies a fluorescently tagged cell through laser emission. FACS can detect the volume and size of cells and sort the cell population of interest from a heterogeneous group of sub-populations.15 Although FACS is a powerful tool it lacks spatial information required for locating a cell within its environment and among other cells.
Newer image-based cytometry techniques (next-generation FACS) utilize laser ablation of metal isotope-tagged antibodies bound to target antigens to provide more spatial information. This tool has been used to detect more than 30 biomarkers on FFPE samples, providing valuable information on the tumor microenvironment and its cellular neighborhoods by positionally identifying appropriate biomarkers. The use of metal tags instead of traditional fluorophores probes also reduces spectral background signaling.16, 17
4. Automation and Resolution
4.1. Automation
The current landscape of spatial technology requires extensive human interference and has relatively low throughput.18,19 However, companies and academic institutes are collaborating to design an end-to-end automated workflow spatial solution. Studies are underway to automate RNA and protein visualization through multiplexing with high sensitivity and specificity.19
4.2. Resolution
Techniques such as matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) and computational algorithms are being tested preclinical to confirm whether proteins/molecules can be identified at a subcellular level with a resolution of around 10 µm. Studies are also being conducted to combine Imaging mass spectrometry such as MALDI and MS with high plex IHC for better and more efficient resolution of cellular content.
In situ capture is another method where sample mRNA binds to the unique spot that contains a specialized barcoded sequence and poly(dT) tail. The poly(dT) tail is hybridized to the corresponding RNA sequence of the tissue sample. After the mRNA is captured, a DNA library is constructed and archived, storing gene-specific information. Depending on the spot size, the DNA library can be resolve/sequence up to 20 cells.8 With the availability of an advanced clustering statistical model, these barcoded transcripts can be further broken down into sub-spots to identify/investigate single cell heterogeneity to the level of 100 nm. 20 An efficient cellular resolution can help differentiate between malignant and non-malignant areas, thereby developing an appropriate treatment regimen for patients in the future.
There are novel methods that are being developed continuously, primarily by combining already developed techniques (scRNAseq, IF, IHC, MALDI, and MS) to gain better resolution of available spatial information.
5. AI Integration
Artificial intelligence (AI) has become an essential tool in spatial biology. AI-trained algorithms and convolutional neural networks help distinguish between disease subtypes and predict an appropriate treatment for a particular disease. AI has developed algorithms to stratify cancer subtypes, including non-small cell lung cancer, and is continuously evolving to improve its diagnostic precision and turnaround time. Organizations are developing algorithms to analyze and capture patient-specific genomic, transcriptomic, and proteomic data.21 These algorithms can collect voluminous data in the form of whole slide images and relay information that coherently defines a problem and directs the end user toward a viable solution. IHC and other assays are used to validate the computer-generated spatial mapping of the corresponding genome, and these validations have proven the accuracy and precision of AI in practice.8
With this wealth of knowledge, clinicians can structure these analyses into actionable clinical information and provide the most appropriate therapeutic regimen to the patient.
6. Future of Spatial Biology
Spatial biology, especially in the last decade, has garnered significant attention.2 With its considerable and growing role in omics analysis, the value of spatial biology is just beginning to be realized by clinicians. Conventionally, cell-based information was gathered in a homogenized tissue to understand RNA or protein levels of the molecules of interest. 2 Spatial biology has made it possible to explore the single cell phenotype in situ, relative to the neighboring cells. Currently, spatial research is focusing on integrating transcriptomics and proteomics so that molecular changes at the level of RNA and protein can be analyzed in tandem and interpreted in the context of disease management.22
Also, AI-based algorithms are being developed that can put together a big picture from several areas of a tissue sample to analyze scattered spatial data points into a continuous piece of actionable information.
Table 2. Advantages and limitations of In-situ Spatial Biology
| Advantage | Limitation |
|---|---|
| Ability to locate cellular and subcellular components | Cannot identify cellular components in live cells |
| Cellular contents can localized in situ; does not require breaking down the tissue into individual cells | Spatial technology to identify entire proteome, transcriptome, or genome in a whole tissue is rare and not a commonly used method |
| Multiplexing helps in the determination of cellular microenvironment | Manual interference makes spatial research time intensive and expensive |
| Promising results in preclinical and some clinical trials | Full clinical adoption is many years away |
Along with its significant advantages, this technology has its own limitations and challenges, as shown in Table 2.
Spatial biology will grow to become more precise and inexpensive for future clinical adaptation and patient benefit. The active use of robotics, AI-trained algorithms, and high throughput requirements is expensive and resource-intensive. In addition, while spatial analysis is currently limited to fixed tissue samples, studies are ongoing to find ways to decipher spatial information in live cells.23
Although it has a long path before being used in the clinic, the future of spatial biology looks promising. Improvements in spatial resolution, advanced computational tools, extraction of substantial information in a reduced turnaround time, and rigorous testing of novel methods and protocols in statistically powered studies are some of the ways to improve the probability of spatial research adaption for clinical use.
7. Conclusion
The field of spatial biology is continuously evolving. The rapid advancement in the area, which includes higher plexing of biological molecules (RNA or protein), automated end-to-end solutions, and successful application of these upgrades in clinical trials, clearly indicates the potential for future growth of spatial biology research and the opportunity this field will provide to improve human lives.
About the presenters

Shubham Dayal is a Senior Medical Writer at Leica Biosystems and has over 10 years of experience in regulatory/preclinical/clinical writing for studies that are at different stages of the product lifecycle. Shubham has a PhD in Cell and Molecular Biology from the University of Toledo and a Master's in Regulatory Affairs from Northeastern University and has co-authored multiple peer-reviewed articles and poster presentations. He is an active member of the Regulatory Affairs Professional Society and American Medical Writers Association and holds certifications related to scientific writing. In his current role, Shubham's goal is to create awareness for our customers in ways that can advance scientific communication and ultimately improve patient outcomes.

Jack obtained his doctorate in molecular and cellular pathology and performed post-doctoral studies on cancer epigenetics and cardiovascular post-translational modifications. He has worked in externally facing roles at Leica Biosystems for 7 years, and currently works in partnership with leading pathologists and researchers to advance scientific study at the cutting edge of anatomic pathology research.
References
- NanoString. An Introduction to Spatial Biology and Spatial Profiling. Accessed September 25, 2023. https://nanostring.com/blog/an-introduction-to-spatial-biology-and-spatial-profiling/
- Tian Yu. Future Trends in Spatial Biology. August 10, 2023. Accessed September 20, 2023. https://thepathologist.com/diagnostics/future-trends-in-spatial-biology
- Akoya Biosciences. Why Spatial Biology. Accessed September 20, 2023.
- Vandereyken, K., Sifrim, A., Thienpont, B. et al. Methods and applications for single-cell and spatial multi-omics. Nat Rev Genet 24, 494–515 (2023). https://doi.org/10.1038/s41576-023-00580-2
- Williams, C.G., Lee, H.J., Asatsuma, T. et al. An introduction to spatial transcriptomics for biomedical research. Genome Med 14, 68 (2022). https://doi.org/10.1186/s13073-022-01075-1
- Lewis SM, Asselin-Labat ML, Nguyen Q, et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat Methods. 2021;18(9):997-1012. doi:10.1038/s41592-021-01203-6
- Lundberg, E., Borner, G.H.H. Spatial proteomics: a powerful discovery tool for cell biology. Nat Rev Mol Cell Biol 20, 285–302 (2019). https://doi.org/10.1038/s41580-018-0094-y
- Masha Savelieff. Spatial Biology: Location, Location, Location! Published February 27, 2023. Accessed 28, September 2023. https://www.technologynetworks.com/proteomics/articles/spatial-biology-location-location-location-370156
- Akoya Biosciences. Spatial Multiomics: The Need for Multidimensional Analysis with Single-Cell, Spatial Context. Accessed 15 September, 2023. https://www.akoyabio.com/blog/spatial-multiomics-the-need-for-multidimensional-analysis-with-single-cell-spatial-context/.
- Locke D, Hoyt CC. Companion diagnostic requirements for spatial biology using multiplex immunofluorescence and multispectral imaging. Front Mol Biosci. 2023;10:1051491. Published 2023 Feb 9. doi:10.3389/fmolb.2023.1051491
- Genetic Engineering and Biotechnology News. Shifting the Paradigm of Modern Spatial Biology. Accessed 10 September 2023. https://www.genengnews.com/insights/shifting-the-paradigm-of-modern-spatial-biology/.
- Lunaphore. Lighten up your spatial biology journey: tips for selecting the perfect secondary antibody for immunofluorescence staining. Published on 7 March 2023. Accessed 10 September 2023. https://lunaphore.com/our-blog/lighten-up-your-spatial-biology-journey-tips-for-selecting-the-perfect-secondary-antibody-for-immunofluorescence-staining/
- nanoString. Common questions in molecular biology: What is DNA barcoding, and why is it important? Published on March 9, 2023. Accessed 20 September 2023. https://nanostring.com/blog/what-is-dna-barcoding-and-why-is-it-important/
- nanoCollect. Flow Cytometry and FACS: What’s the Difference? Published on 17 February 2020. Accessed September 1, 2023. https://nanocellect.com/blog/flow-cytometry-vs-facs/
- Elaldi R, Hemon P, Petti L, et al. High Dimensional Imaging Mass Cytometry Panel to Visualize the Tumor Immune Microenvironment Contexture. Front Immunol. 2021;12:666233. Published 2021 Apr 16. doi:10.3389/fimmu.2021.666233
- Kay AW, Strauss-Albee DM, Blish CA. Application of Mass Cytometry (CyTOF) for Functional and Phenotypic Analysis of Natural Killer Cells. Methods Mol Biol. 2016;1441:13-26. doi:10.1007/978-1-4939-3684-7_2
- Piehowski, P.D., Zhu, Y., Bramer, L.M. et al. Automated mass spectrometry imaging of over 2000 proteins from tissue sections at 100-μm spatial resolution. Nat Commun 11, 8 (2020). https://doi.org/10.1038/s41467-019-13858-z
- Berglund, E., Saarenpää, S., Jemt, A. et al. Automation of Spatial Transcriptomics library preparation to enable rapid and robust insights into spatial organization of tissues. BMC Genomics 21, 298 (2020). https://doi.org/10.1186/s12864-020-6631-z
- Medical Device Network. Bio-Techne and Lunaphore to develop automated spatial multi-omics solution. Published April 18, 2023. Accessed 1 September 2023. https://www.medicaldevice-network.com/news/bio-techne-lunaphore-automated-spatial-multiomics-solution/
- Vizgen. What is Spatial Genomics? Published November 4, 2022. Accessed 25 September 2023. https://vizgen.com/what-is-spatial-genomics/
- Genetic Engineering and Biotechnology News. Spatial Biology and AI Combined by Noetik to Develop Immuno-Oncology Drugs. Published September 11, 2023. Accessed September 15 2023. https://www.genengnews.com/topics/drug-discovery/spatial-biology-and-ai-combined-by-noetik-to-develop-immuno-oncology-drugs/
- Artificial intelligence-powered spatial biology is transforming precision medicine. Nature. Accessed September 02, 2023. https://www.nature.com/articles/d43747-022-00124-3
- Joanna Owens. Spatial Biology: The Next Revolution in Understanding Health and Disease. Published November 04, 2022. Accessed August 15 2023. https://www.technologynetworks.com/proteomics/articles/spatial-biology-the-next-revolution-in-understanding-health-and-disease-367236
Related Content
Leica Biosystems content is subject to the Leica Biosystems website terms of use, available at: Legal Notice. The content, including webinars, training presentations and related materials is intended to provide general information regarding particular subjects of interest to health care professionals and is not intended to be, and should not be construed as, medical, regulatory or legal advice. The views and opinions expressed in any third-party content reflect the personal views and opinions of the speaker(s)/author(s) and do not necessarily represent or reflect the views or opinions of Leica Biosystems, its employees or agents. Any links contained in the content which provides access to third party resources or content is provided for convenience only.
For the use of any product, the applicable product documentation, including information guides, inserts and operation manuals should be consulted.
Copyright © 2025 Leica Biosystems division of Leica Microsystems, Inc. and its Leica Biosystems affiliates. All rights reserved. LEICA and the Leica Logo are registered trademarks of Leica Microsystems IR GmbH.


