
Protease-Free Workflow for Spatial Multiomics Analysis of the Tumor Microenvironment in Breast Cancer
Contributors
Tanni Rahman, PhD1, Patrick J. Savickas1, Rachel C. Clary, PhD1, Andrzej Cholewinski, PhD1, Anushka Dikshit, PhDs, Joseph Iacona, PhD2, Kristy Chu2
1HistoWiz, New York, NY
2Advanced Cell Diagnostics, a Bio-Techne brand, Newark, CA
Introduction
Breast cancer is the most commonly diagnosed cancer among women worldwide1. Notably, 1 in 8 women will develop invasive breast cancer during their lifetime1. Continued research of the biology underlying breast cancer is essential to the development of novel therapeutics and biomarker detection methods.
Multiomic spatial resolution of the tumor microenvironment
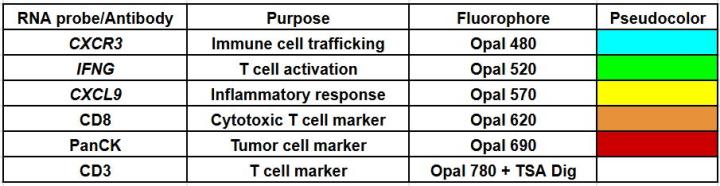
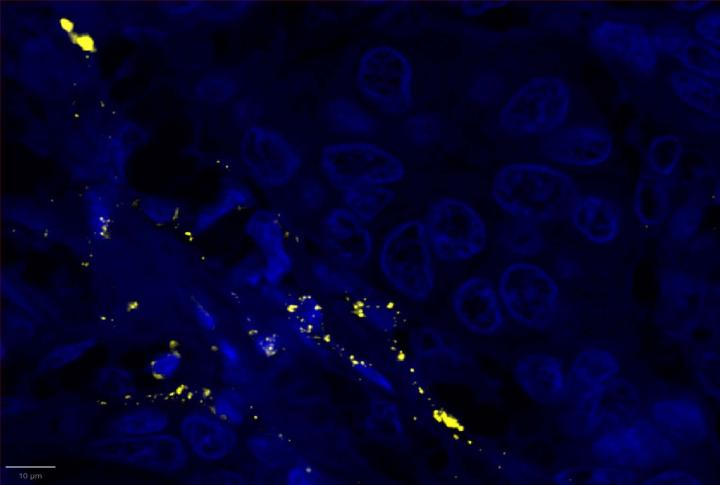
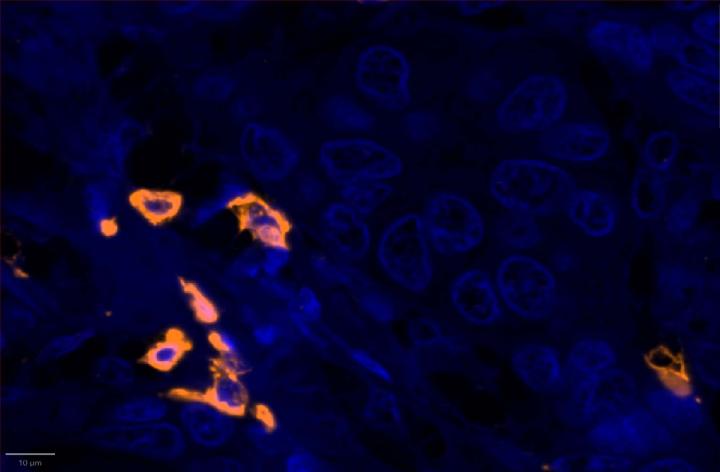
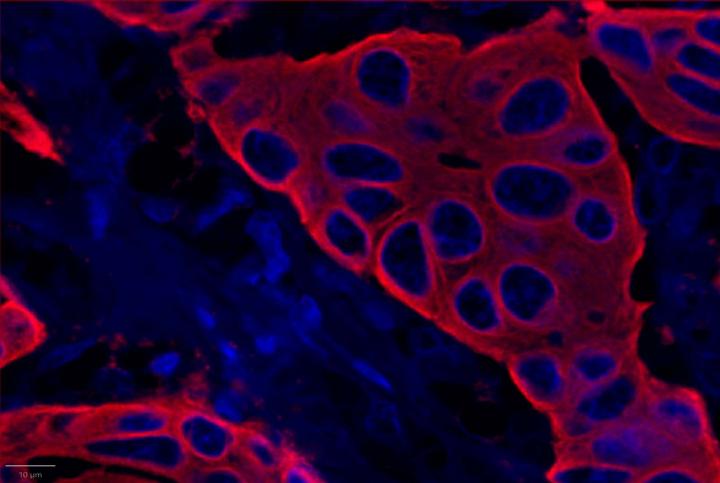
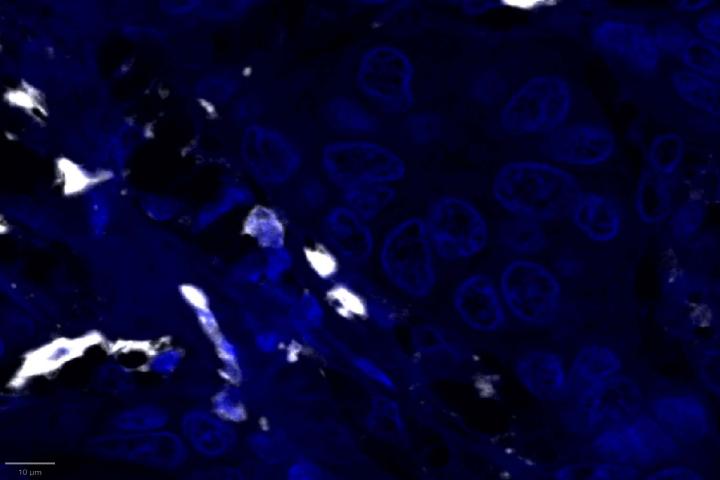
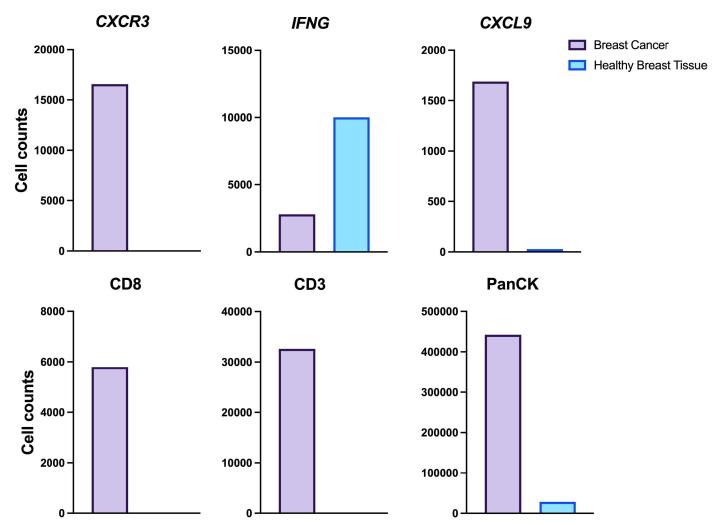
Here, we utilized RNAscope™ Multiomic LS technology to stain RNA and protein markers, CXCR3, IFNG, CXCL9, CD3, PanCK, and CD8. CXCR3 is indicative of immune cell trafficking, IFNG marks T cell activation, and CXCL9 is involved in inflammatory responses relevant to anti-tumor immunity 2-4. In conjunction, CD3 and CD8 are protein markers for T cells, and PanCK delineates tumor positive areas 5-6. Together, we have utilized a novel protease-free workflow to simultaneously detect RNA and protein markers in breast cancer tissues to further understand the tumor microenvironment.
Materials and Methods
- All stains were performed using the BOND RX fully automated research stainer
- Formalin-fixed, paraffin-embedded human breast cancer and healthy breast tissue sections were used for multiomic staining.
For Research Use Only. Not for use in diagnostic procedures.

RNAscope™ Multiomic LS Assay Schematic
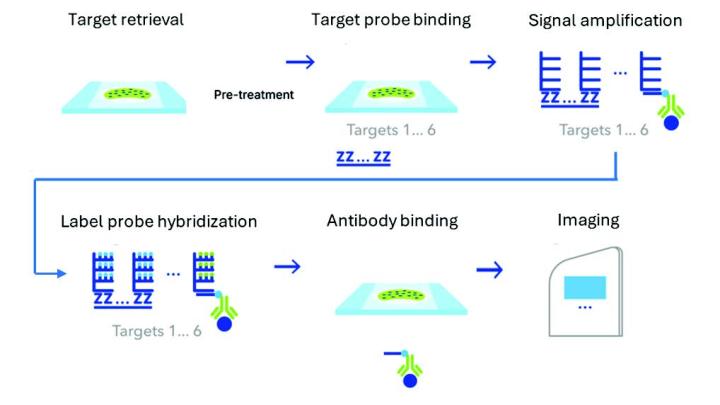
- Samples were imaged at 40X magnification, followed by image analysis.
- The new Multiomic LS assay supports detection of 6-plex RNA+protein targets with TSA-based detection. The workflow, run on the BOND RX, involves protease-free target retrieval followed by probe binding, signal amplification, and detection. This was followed by antibody binding for protein detection.
RNA Target, Antibody, and Fluorophore Pairings
Staining Results
Positive Control Multiomic Staining in Breast Cancer
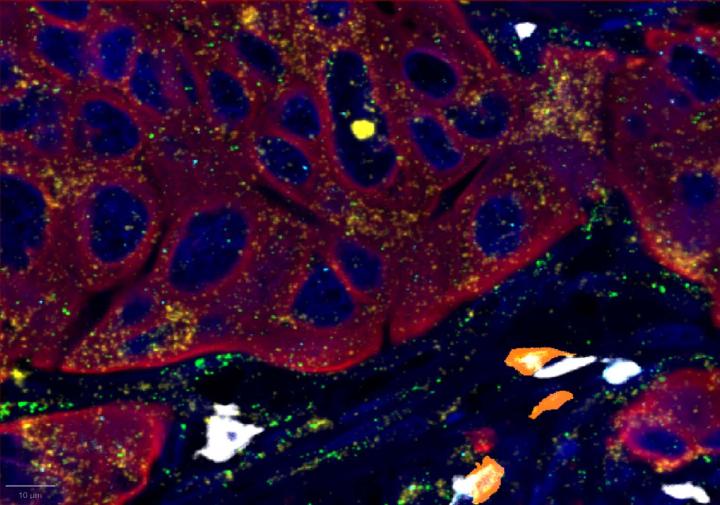
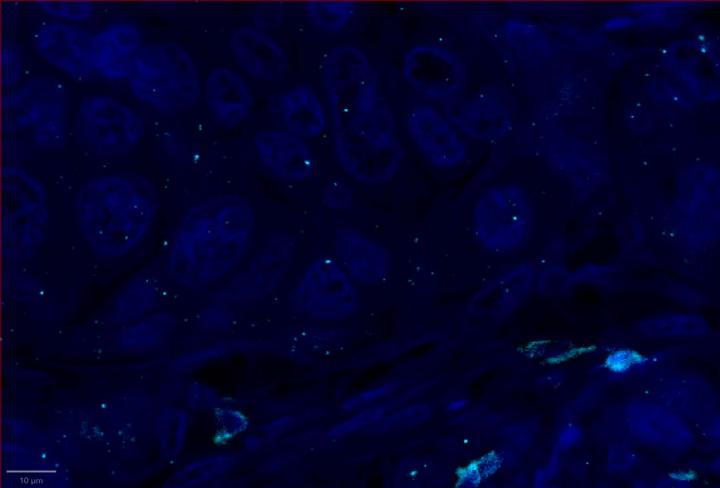
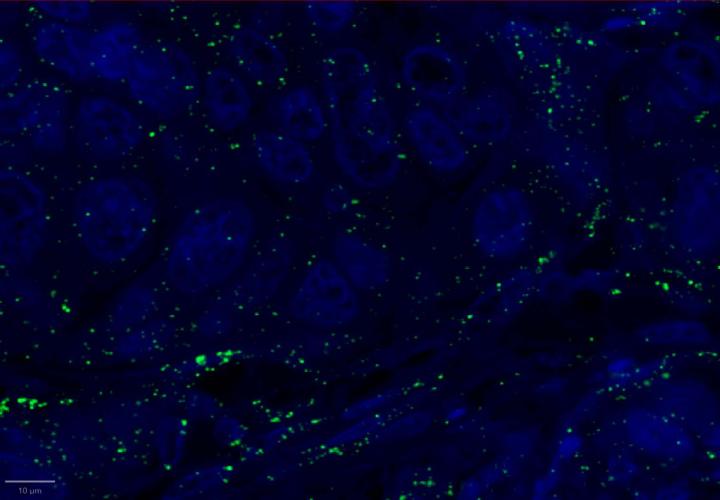
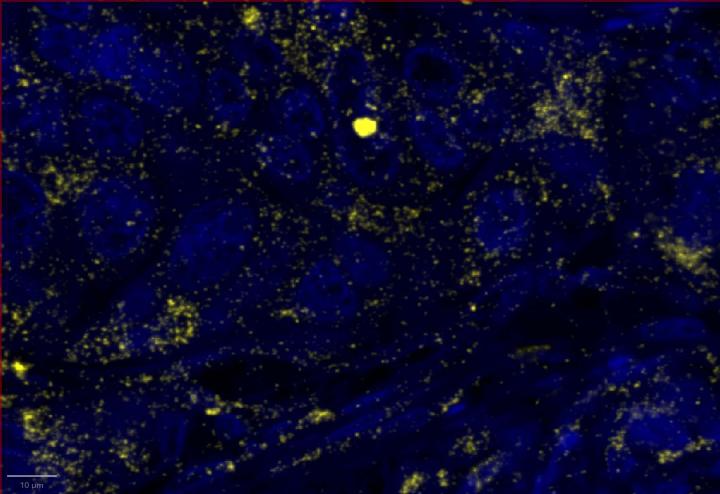
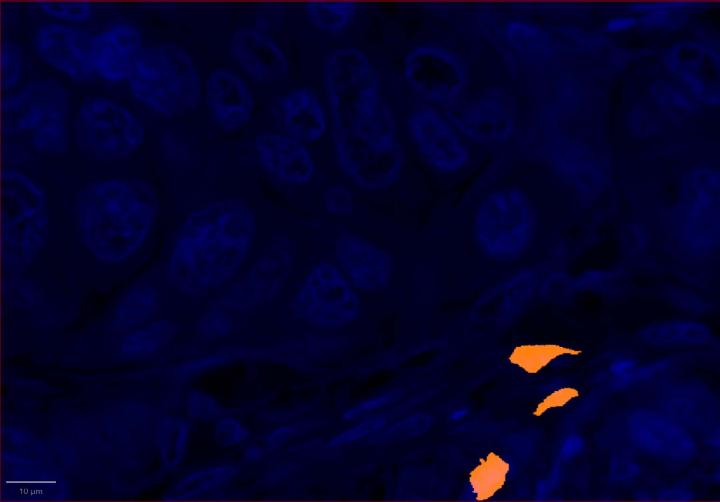
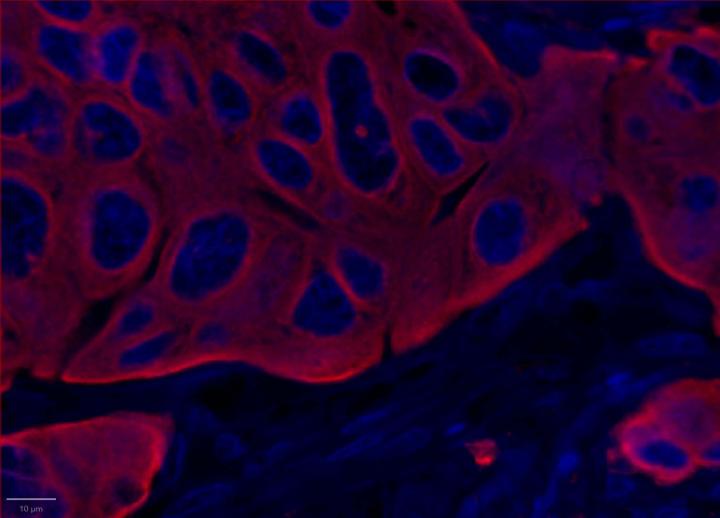
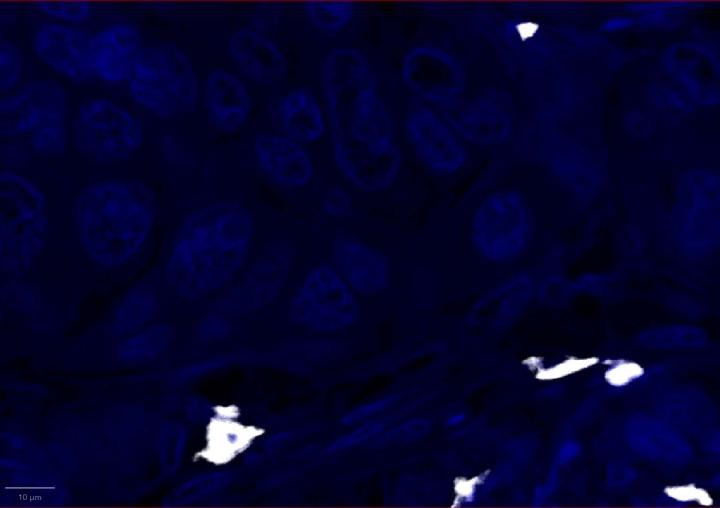
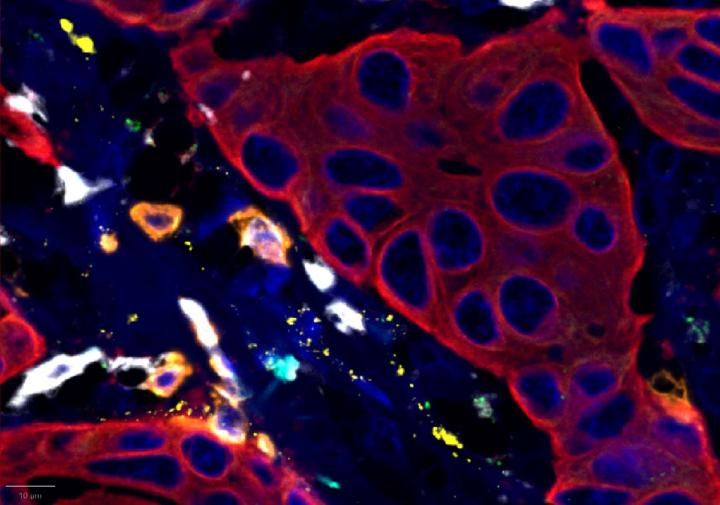
Experimental Multiomic Staining in Breast Cancer
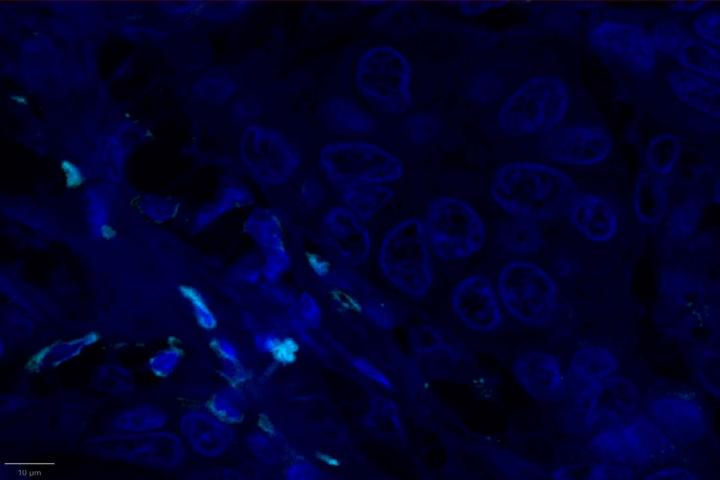
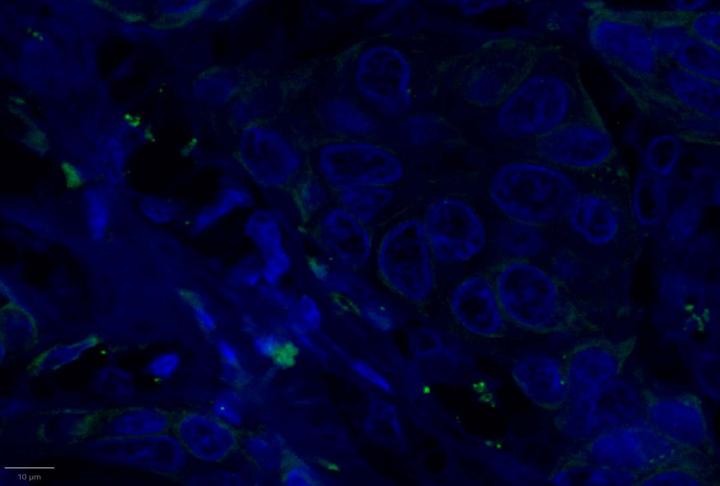
AI-Driven Image Analysis
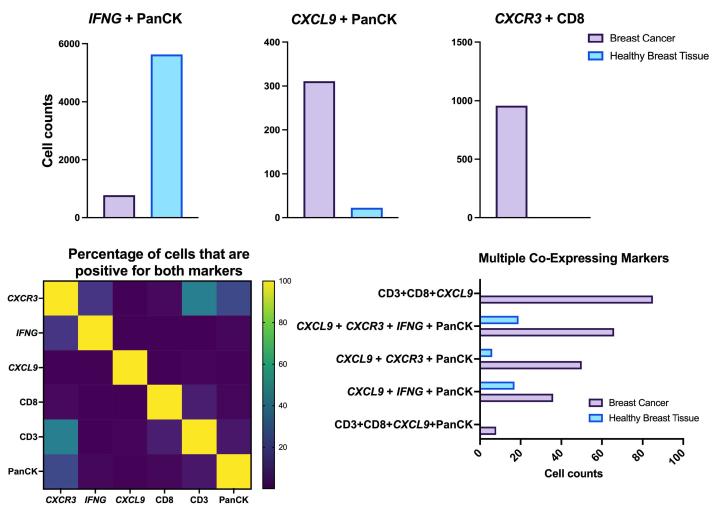
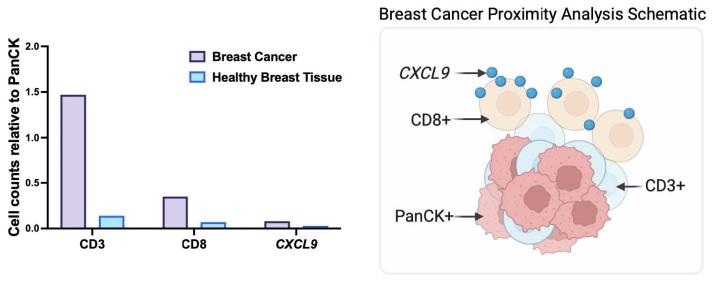
Proximity Analysis Results
Conclusions
- Compared to healthy breast tissue, the expression of CXCR3, CXCL9, CD3, CD8, and PanCK was elevated in breast cancer. This suggests strong immune cell infiltration within the tumor microenvironment. Elevated CXCR3 expression is associated with enhanced immune surveillance, while increased CXCL9 may reflect active anti-tumor immunity2,4.
- Multiomic analysis revealed co-expression of CXCR3 with CD8, indicative of T-cell recruitment7. IFNG was co-expressed with PanCK, but was down-regulated in breast cancer relative to healthy tissue, potentially indicating immune suppression and poor prognosis8.
- Proximity analysis revealed that in breast cancer, CD3, CD8, and CXCL9 were spatially adjacent to PanCK+ tumor cells, suggesting a localized pro-inflammatory response driven by cytotoxic T-cell activity9.
- The RNAscope™ Multiomic LS technology provides a powerful tool to deepen our understanding of breast cancer biology and the immune landscape of the tumor microenvironment.
Want to learn how the BOND RX can support cancer research in your lab?
References
- Giaquinto, A. N., Sung, H., Newman, L. A., Freedman, R. A., Smith, R. A., Star, J., Jemal, A., & Siegel, R. L. (2024). Breast cancer statistics, 2024. CA: A Cancer Journal for Clinicians, 74(6), 477–495. https://doi.org/10.3322/caac.21863
- Groom, J. R., & Luster, A. D. (2011). CXCR3 in T cell function. Experimental cell research, 317(5), 620–631. https://doi.org/10.1016/j.yexcr.2010.12.017
- Jorgovanovic, D., Song, M., Wang, L., Zhang, Y. Roles of IFN-γ in tumor progression and regression: a review. Biomark Res 8, 49 (2020). https://doi.org/10.1186/s40364-020-00228-x
- Tokunaga, R., Zhang, W., Naseem, M., Puccini, A., Berger, M. D., Soni, S., McSkane, M., Baba, H., & Lenz, H. J. (2018). CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - A target for novel cancer therapy. Cancer treatment reviews, 63, 40–47. https://doi.org/10.1016/j.ctrv.2017.11.007
- Sun X, Zhai J, Sun B, Parra ER, Jiang M, Ma W, Wang J, Kang AM, Kannan K, Pandurengan R, Zhang S, Solis LM, Haymaker CL, Raso MG, Mendoza Perez J, Sahin AA, Wistuba II, Yam C, Litton JK, Yang F. Effector memory cytotoxic CD3+/CD8+/CD45RO+ T cells are predictive of good survival and a lower risk of recurrence in triple-negative breast cancer. Mod Pathol. 2022 May;35(5):601-608. doi: 10.1038/s41379-021-00973-w. Epub 2021 Nov 27. PMID: 34839351.
- Ji H, Yuan L, Jiang Y, Ye M, Liu Z, Xia X, Qin C, Jiang D, Gai Y, Lan X. Visualizing Cytokeratin-14 Levels in Basal-Like Breast Cancer via ImmunoSPECT Imaging. Mol Pharm. 2022 Oct 3;19(10):3542-3550. doi: 10.1021/acs.molpharmaceut.2c00004. Epub 2022 Mar 14. PMID: 35285645.
- Kurachi, M., Kurachi, J., Suenaga, F., Tsukui, T., Abe, J., Ueha, S., Tomura, M., Sugihara, K., Takamura, S., Kakimi, K., & Matsushima, K. (2011). Chemokine receptor CXCR3 facilitates CD8(+) T cell differentiation into short-lived effector cells leading to memory degeneration. The Journal of experimental medicine, 208(8), 1605–1620. https://doi.org/10.1084/jem.20102101
- Lamsal, A., Andersen, S. B., Johansson, I., Vietri, M., Bokil, A. A., Kurganovs, N. J., Rylander, F., Bjørkøy, G., Pettersen, K., & Giambelluca, M. S. (2023). Opposite and dynamic regulation of the interferon response in metastatic and non-metastatic breast cancer. Cell communication and signaling : CCS, 21(1), 50. https://doi.org/10.1186/s12964-023-01062-y
- Liang, H., Huang, J., Li, H., He, W., Ao, X., Xie, Z., Chen, Y., Lv, Z., Zhang, L., Zhong, Y., Tan, X., Han, G., Zhou, J., Qiu, N., Jiang, M., Xia, H., Zhan, Y., Jiao, L., Ma, J., Radisky, D., … Zhang, X. (2025). Spatial proximity of CD8+ T cells to tumor cells predicts neoadjuvant therapy efficacy in breast cancer. NPJ breast cancer, 11(1), 13. https://doi.org/10.1038/s41523-025-00728-9
Related Content
Leica Biosystems content is subject to the Leica Biosystems website terms of use, available at: Legal Notice. The content, including webinars, training presentations and related materials is intended to provide general information regarding particular subjects of interest to health care professionals and is not intended to be, and should not be construed as, medical, regulatory or legal advice. The views and opinions expressed in any third-party content reflect the personal views and opinions of the speaker(s)/author(s) and do not necessarily represent or reflect the views or opinions of Leica Biosystems, its employees or agents. Any links contained in the content which provides access to third party resources or content is provided for convenience only.
For the use of any product, the applicable product documentation, including information guides, inserts and operation manuals should be consulted.
Copyright © 2026 Leica Biosystems division of Leica Microsystems, Inc. and its Leica Biosystems affiliates. All rights reserved. LEICA and the Leica Logo are registered trademarks of Leica Microsystems IR GmbH.




